Pinterest's Embedding Based Retrieval
Google's AI Co-Scientist
Pinterest’s wrote a blog post on their retrieval system to enhance personalization, engagement, and diversity through innovations in embedding-based retrieval and conditional retrieval. I covered the learned retrieval system in the previous post here:
This post is post followed to the above post where Pinterest showcases how they have improved upon the embedding based retrieval and implemented a conditional retrieval.
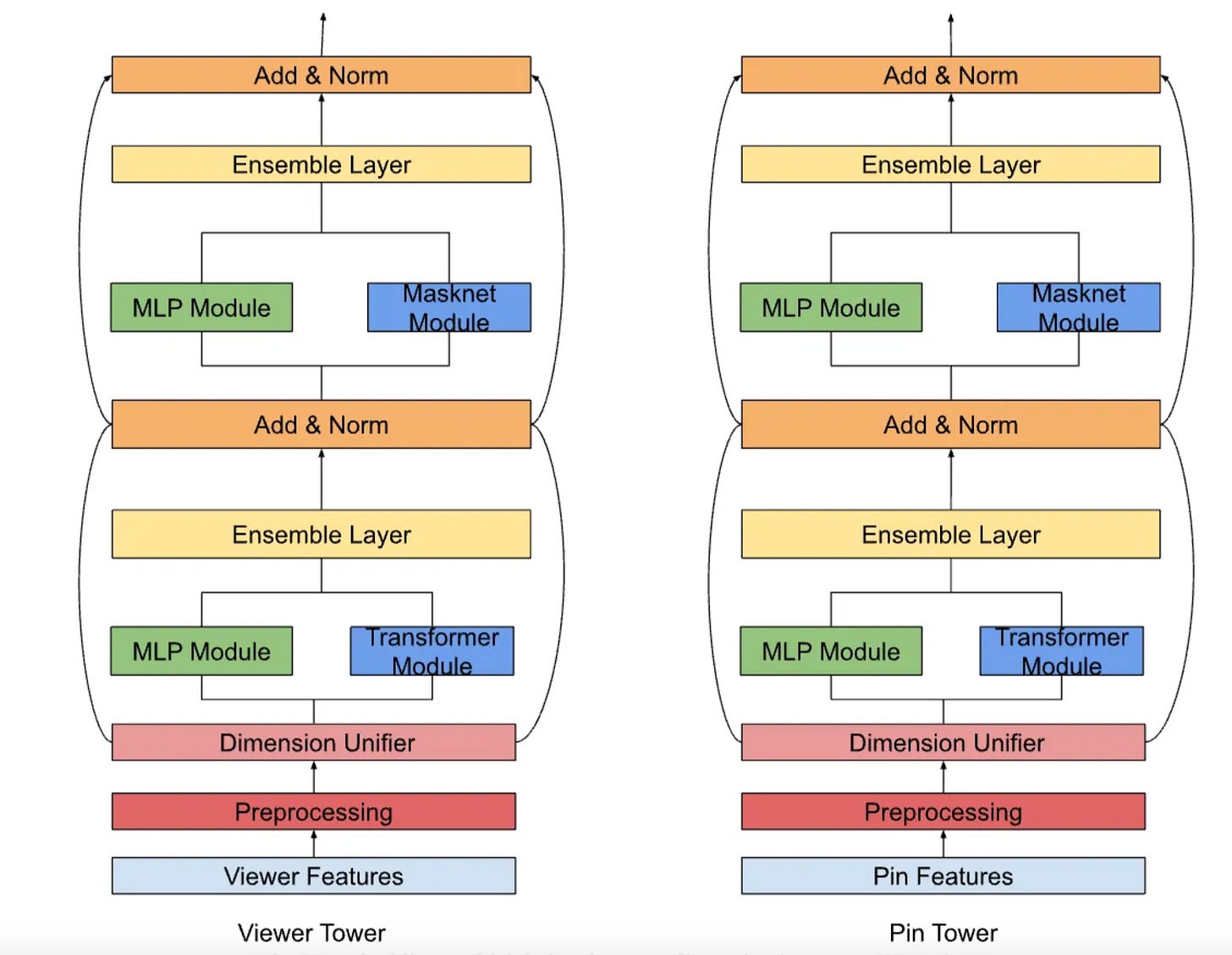
Pinterest’s retrieval system is a two-tower neural network comprising a user tower and an item (Pin) tower. The user tower processes real-time engagement sequences (e.g., clicks, saves) and contextual signals(e.g., device type, location), while the Pin tower encodes visual features(CNN-extracted embeddings), textual metadata(BERT embeddings), and statistical features(e.g., historical engagement rates). Both towers output dense embeddings trained via contrastive learning with in-batch negative sampling and a debiased loss function, optimizing dot-product similarity between user and Pin representations.
To address the inherent limitation of two-tower models—no direct user-item feature interaction—Pinterest introduced bitwise and field-wise feature crossing within each tower:
MaskNet Integration: MaskNet blocks apply Hadamard products between input embeddings and MLP-projected representations, followed by refinement layers. Parallelizing four MaskNet blocks with bottleneck MLPs improved engaged sessions (sessions >60 seconds) by 0.15–0.35%.
DHEN Framework: The Deep and Hierarchical Ensemble Network (DHEN) combines MaskNet with transformer encoders for field-level attention, enabling juxtaposed MLP and transformer layers. This hybrid approach boosted engagement metrics by +0.1–0.2% and homefeed saves by >1%.
To mitigate ID embedding overfitting, Pinterest pre-trains user and Pin ID embeddings via contrastive learning on a cross-surface dataset spanning multiple engagement types (e.g., saves, clicks). Some of the technical approaches they adopted while doing so:
Non-overlapping Training Windows: Using pretrained IDs from periods outside the fine-tuning window to reduce overfitting, yielding a 0.25–0.35% increase in repins.
Aggressive Dropout: Applying 50% dropout on ID embeddings during fine-tuning, which improved homefeed repins and clicks by 0.6–1.2%.
Distributed Training: Leveraging torchrec library for GPU-shared embedding tables, enabling efficient handling of billion-scale ID spaces.
Despite training on GPUs, the final model serves CPU-based embeddings due to latency requirements for offline Pin tower pre-computation. This design choice balances precision with Pinterest’s need for 50ms p99 latency during real-time user embedding inference.
The serving corpus—the set of retrievable Pins—was upgraded from a static 90-day engagement sum to a time-decayed scoring system:
Score(p,d)=∑iengagementi⋅e−λ(d−di)
where λ controls decay rate. This prioritizes recent trends, improving retrieval relevance and aligning training/serving data distributions.
A granularity mismatch between training (fine-grained image signatures) and serving (coarse deduplicated signatures) caused feature drift. Pinterest implemented a remapping layer to reconcile these differences, achieving a +0.1–0.2% engaged session lift without model retraining.
Capsule Network Adaptation
To capture diverse user intents, Pinterest replaced single-embedding user representations with multi-embedding clusters using a modified Capsule Network. Key changes that they have adopted:
MaxMin Initialization: Accelerating cluster convergence by maximizing initial centroid distances.
Single-Assignment Routing: Restricting user history items to contribute to only one cluster, enhancing diversity.
At serving time, the top-K clusters (determined by user history length) undergo ANN search via HNSW graphs, with results blended in round-robin fashion. This increased Pin save actions by better aligning with users’ exploratory intents.
Conditional Retrieval for Interest-Driven Personalization
Pinterest’s conditional retrieval system augments the two-tower model with auxiliary interest signals (explicit follows + inferred interests).
During training:
The user tower receives a concatenated input of engagement history and a target Pin’s interest ID (early fusion).
The model learns to associate interest-conditioned embeddings with relevant Pins, even for long-tail interests.
At inference, ANN searches are filtered by interest ID matches, ensuring retrieved Pins align with the queried interest. This hybrid approach improved recommendation funnel efficiency by balancing personalization and diversity.
Google Research's published a blog post on their AI co-scientist system represents a large shift in human-AI collaboration for scientific advancement.
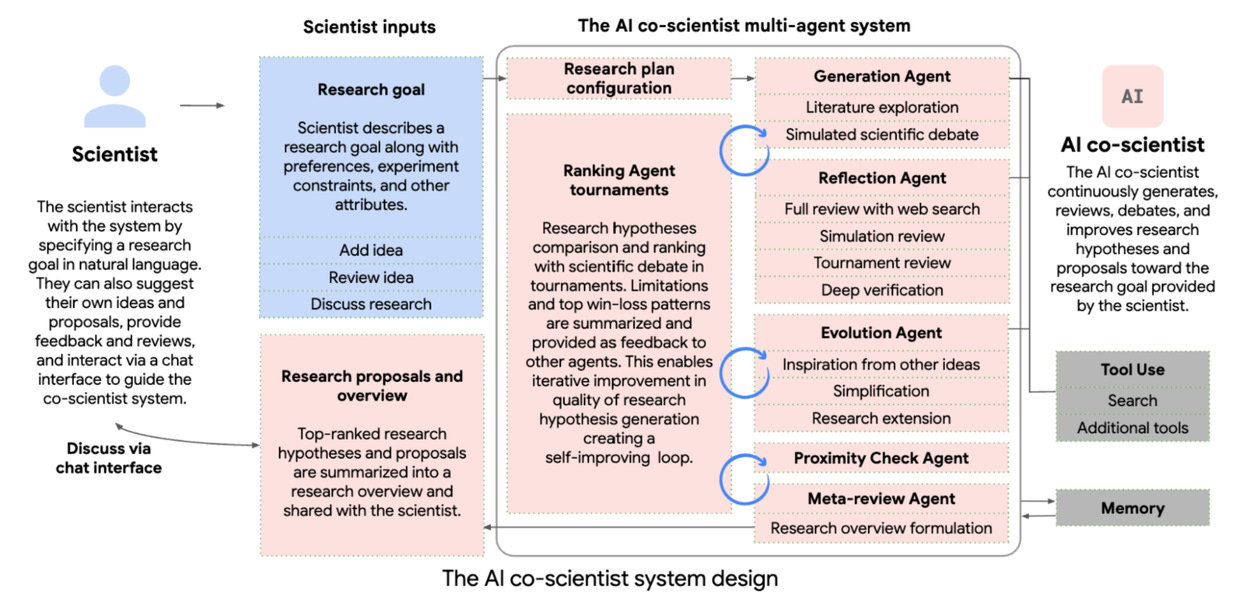
Fundamentally, the AI co-scientist system represents an improved implementation of multi-agent LLM network built upon the Gemini 2.0 framework. Unlike conventional literature review tools, this system employs a dynamic coalition of specialized agents modeled after key components of the scientific method itself. This is pretty interesting as the scientific method is mapped to different agents directly and agents’ optimization outcomes specifically correlate with these outputs.
The architectural design features six primary agent types(completely adopting agents framework):
Generation Agent: Responsible for initial hypothesis formulation through combinatorial analysis of existing scientific knowledge
Reflection Agent: Conducts critical evaluation of generated hypotheses through simulated peer review processes
Ranking Agent: Implements tournament-style comparisons between competing hypotheses using Elo rating systems
Evolution Agent: Drives iterative hypothesis refinement through recursive self-improvement cycles
Proximity Agent: Maintains conceptual coherence with original research objectives
Meta-review Agent: Oversees higher-order integration of feedback across the system
A Supervisor agent orchestrates resource allocation and workflow management, enabling dynamic scaling of computational resources based on problem complexity. This hierarchical structure allows the system to simulate human-like scientific reasoning while leveraging machine-speed processing capabilities.
The system's most significant capabilities lies in its ability to generate novel research hypotheses rather than simply synthesizing existing knowledge, which has been the bottleneck problem in generating new information that enables research and innovations. Through a combination of cross-domain knowledge integration and self-play debate mechanisms, the AI co-scientist demonstrates capacity for producing testable predictions that extend beyond current scientific consensus.
The innovations that enabled this capability:
Test-Time Compute Scaling: The system dedicates variable computational resources based on problem difficulty, allowing deeper exploration of complex scientific questions.
Automated Feedback Loops: Hypotheses undergo multiple refinement cycles combining synthetic peer review and empirical validation against existing datasets.
Transdisciplinary Synthesis: By integrating concepts from disparate scientific domains, the system mimics the cross-pollination of ideas that drives major breakthroughs in human-led research.
An Elo-style rating system provides quantitative assessment of hypothesis quality, with higher-ranked proposals demonstrating strong correlation with expert assessments on the GPQA benchmark (a standardized measure of scientific reasoning accuracy). In controlled comparisons, the system's outputs were preferred by domain experts over both unaided human proposals and existing AI models in 73% of cases.
Validation Through Experimental Partnerships
Google team conducted validation across three distinct biomedical challenges, partnering with leading academic institutions to assess real-world applicability:
Acute Myeloid Leukemia Drug Repurposing
Working with clinical oncologists and computational biologists, the system identified novel therapeutic applications for existing pharmaceuticals. Subsequent in vitro testing confirmed potent anti-leukemic activity of proposed drug candidates at clinically achievable concentrations. This demonstration of viable drug repurposing highlights the system's ability to navigate complex pharmacokinetic and pharmacodynamic relationships.
Liver Fibrosis Target Discovery
In collaboration with Stanford researchers, the AI co-scientist proposed epigenetic targets demonstrating significant anti-fibrotic activity in human hepatic organoid models. The system's capacity to integrate single-cell sequencing data with pathway analysis enabled identification of previously overlooked regulatory mechanisms in extracellular matrix remodeling.
Antimicrobial Resistance Mechanisms
Partnering with the Fleming Initiative, researchers challenged the system to elucidate horizontal gene transfer mechanisms in bacterial antibiotic resistance. The AI co-scientist independently rediscovered capsid-forming phage-inducible chromosomal islands (cf-PICIs) as key mediators of inter-species resistance gene transfer, matching findings from ongoing (unpublished) laboratory investigations.
Performance Benchmarks and Limitations
System has some good benchmarking results:
The system demonstrated 48% higher hypothesis generation velocity compared to unaided researchers
Proposed hypotheses showed 62% concordance with experimental validation results versus 54% for human-generated proposals
Median time from hypothesis generation to experimental confirmation was reduced by 33% in collaborative workflows
However, it has also a number of limitations:
Domain Specificity: Performance varies significantly across scientific disciplines, with strongest results in biomedicine
Conceptual Drift: Extended reasoning chains occasionally exhibit reduced coherence in highly novel research directions
Validation Dependency: Ultimate hypothesis verification remains dependent on human-led experimental protocols
Libraries
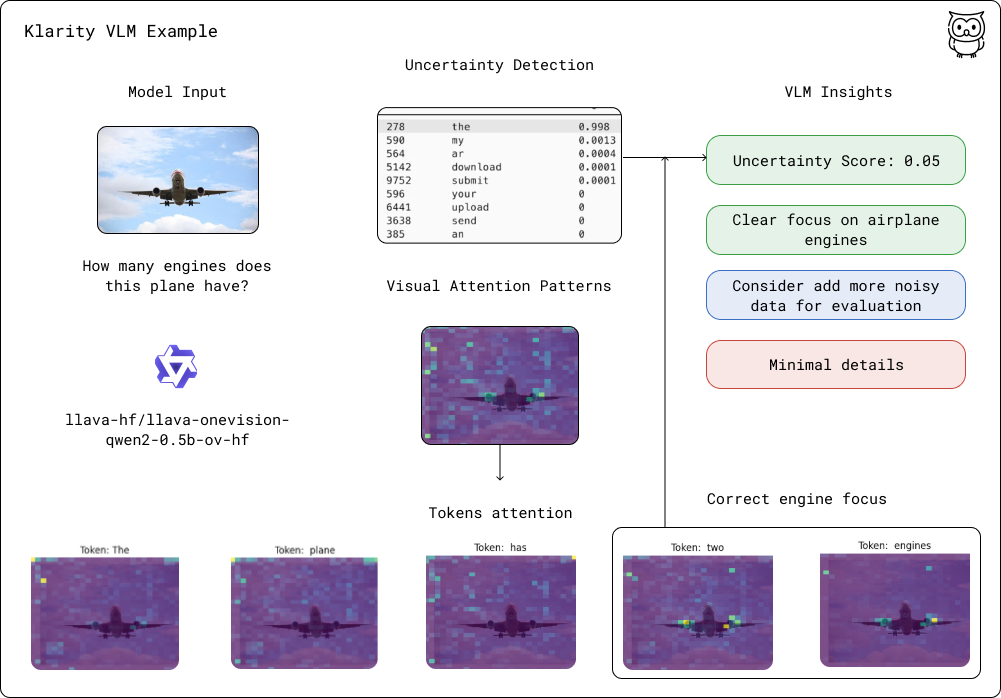
Klarity is a toolkit for inspecting and debugging AI decision-making processes. By combining uncertainty analysis with reasoning insights and visual attention patterns, it helps you understand how models think and fix issues before they reach production.
Dual Entropy Analysis: Measure model confidence through raw entropy and semantic similarity metrics
Reasoning Analysis: Extract and evaluate step-by-step thinking patterns in model outputs
Visual Attention Analysis: Visualize and analyze how vision-language models attend to images
Semantic Clustering: Group similar predictions to reveal decision-making pathways
Structured Insights: Get detailed JSON analysis of both uncertainty patterns and reasoning steps
AI-powered Report: Leverage capable models to interpret generation patterns and provide human-readable insights
ROCm is an open-source stack, composed primarily of open-source software, designed for graphics processing unit (GPU) computation. ROCm consists of a collection of drivers, development tools, and APIs that enable GPU programming from low-level kernel to end-user applications.
With ROCm, you can customize your GPU software to meet your specific needs. You can develop, collaborate, test, and deploy your applications in a free, open source, integrated, and secure software ecosystem. ROCm is particularly well-suited to GPU-accelerated high-performance computing (HPC), artificial intelligence (AI), scientific computing, and computer aided design (CAD).
Open CUA Kit (Computer Use Agent), or Open-CUAK (pronounced "quack" 🦆🗣️), is THE platform for managing automation agents at scale — starting with browsers. The Kubernetes for CUA agents.
🎯 Why Open-CUAK?
In the real world, for real businesses, working with real people, reliability is everything. When automation becomes reliable, it becomes scalable. And when it becomes scalable, it becomes profitable.
That’s why Open-CUAK is designed to run and manage thousands of automation agents, ensuring each one is reliable.
This project is still in its very early days, but our team is working very hard to make it a reality, soon. This is just the beginning of a new era in work, a new way to a world of abundant productivity.
And when productivity becomes truly abundant, we want to make sure it is equally distributed.
bitnet.cpp is the official inference framework for 1-bit LLMs (e.g., BitNet b1.58). It offers a suite of optimized kernels, that support fast and lossless inference of 1.58-bit models on CPU (with NPU and GPU support coming next).
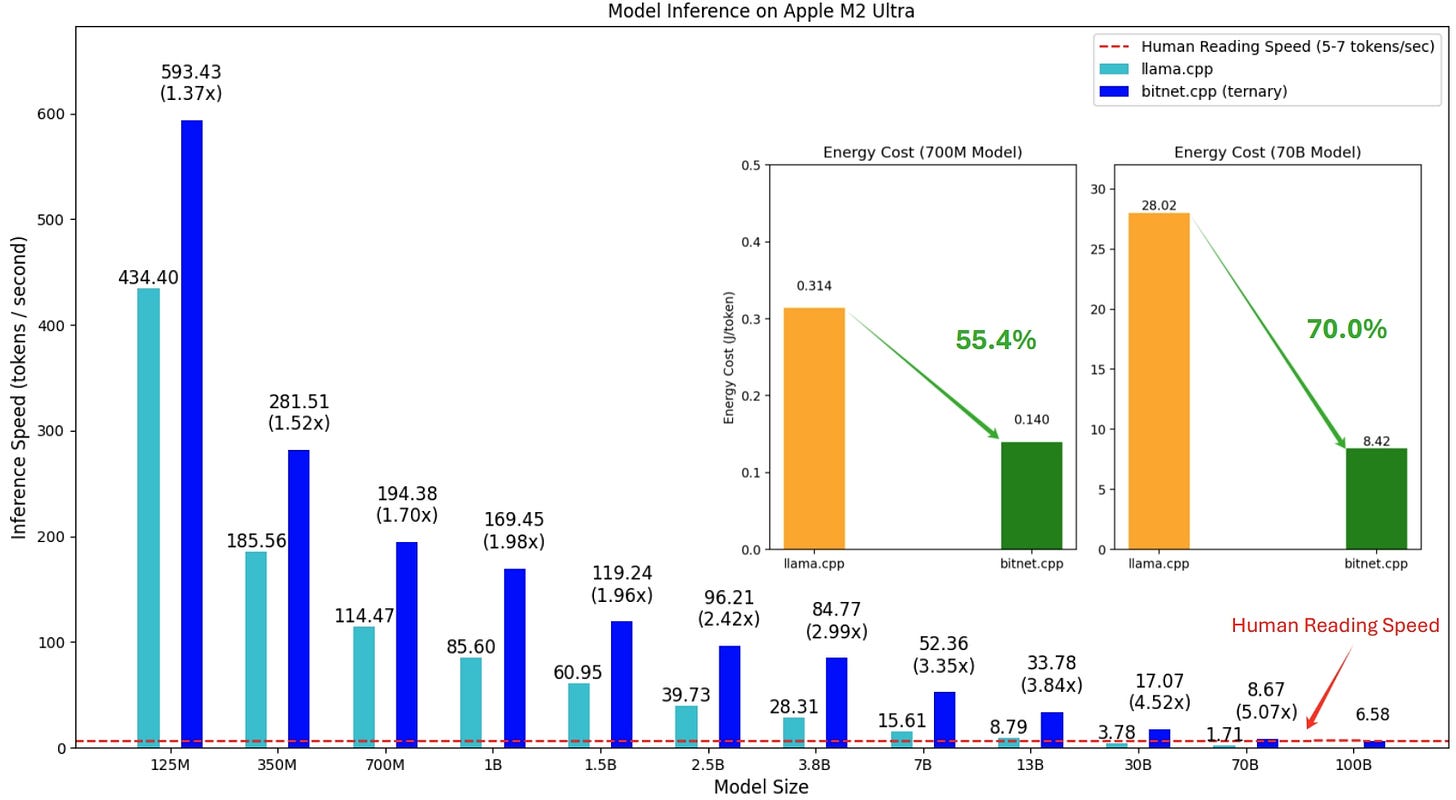
The first release of bitnet.cpp is to support inference on CPUs. bitnet.cpp achieves speedups of 1.37x to 5.07x on ARM CPUs, with larger models experiencing greater performance gains. Additionally, it reduces energy consumption by 55.4% to 70.0%, further boosting overall efficiency. On x86 CPUs, speedups range from 2.37x to 6.17x with energy reductions between 71.9% to 82.2%. Furthermore, bitnet.cpp can run a 100B BitNet b1.58 model on a single CPU, achieving speeds comparable to human reading (5-7 tokens per second), significantly enhancing the potential for running LLMs on local devices. Please refer to the technical report for more details.
LLM Functions empowers you to effortlessly build powerful LLM tools and agents using familiar languages like Bash, JavaScript, and Python.
Forget complex integrations, harness the power of function calling to connect your LLMs directly to custom code and unlock a world of possibilities. Execute system commands, process data, interact with APIs – the only limit is your imagination.
Below The Fold
Earthstar is a small and resilient distributed storage protocol designed with a strong focus on simplicity, and the social realities of peer-to-peer computing kept in mind.
Earthstar is an offline-first database. Earthstar stores all data directly on your device, so that reading and writing works even without an internet connection.
Earthstar is a key-value database which supports author versions. Earthstar databases store arbitrary data, keyed by two pieces of information: a free-form path and a public key identifying the author. So a single path like
/wiki/flowersmay hold revisions by multiple authors.Earthstar data is mutable. Earthstar lets you delete or overwrite the data you have created, but no-one else can.
Earthstar databases can sync. Earthstar databases can by synchronised so that changes from many users and devices can be combined into a reconciled state.
Earthstar databases are private. Earthstar databases do not announce their presence to the world. Only people who know a special identifier can learn of their existence from another peer and replicate their data.
Write access is restricted by a second identifier, so it's possible to grant read-only access to a database.
Earthstar works in the browser. The JavaScript implementation of Earthstar has been designed with standard Web APIs and a pluggable architecture which lets it use APIs exclusive to certain environments, one of them being browsers.
yek is a fast Rust based tool to serialize text-based files in a repository or directory for LLM consumption.
By default:
Uses
.gitignorerules to skip unwanted files.Uses the Git history to infer what files are more important.
Infers additional ignore patterns (binary, large, etc.).
Automatically detects if output is being piped and streams content instead of writing to files.
Supports processing multiple directories in a single command.
Configurable via a
yek.yamlfile.
ChartDB is a powerful, web-based database diagramming editor. Instantly visualize your database schema with a single "Smart Query." Customize diagrams, export SQL scripts, and access all features—no account required. Experience seamless database design here.
What it does:
Instant Schema Import Run a single query to instantly retrieve your database schema as JSON. This makes it incredibly fast to visualize your database schema, whether for documentation, team discussions, or simply understanding your data better.
AI-Powered Export for Easy Migration Our AI-driven export feature allows you to generate the DDL script in the dialect of your choice. Whether you’re migrating from MySQL to PostgreSQL or from SQLite to MariaDB, ChartDB simplifies the process by providing the necessary scripts tailored to your target database.
Interactive Editing Fine-tune your database schema using our intuitive editor. Easily make adjustments or annotations to better visualize complex structures.